Molecular Obstacles to Clinical Translation of iPSCs
The ability to reprogram somatic cells into induced pluripotent stem cells (iPSCs) using defined factors provides new tools for biomedical research. However, some iPSC clones display tumorigenic and immunogenic potential, thus raising concerns about their utility and safety in the clinical setting.
The ability to reprogram somatic cells into induced pluripotent stem cells (iPSCs) using defined factors provides new tools for biomedical research. However, some iPSC clones display tumorigenic and immunogenic potential, thus raising concerns about their utility and safety in the clinical setting. Furthermore, variability in iPSC differentiation potential has also been described. Here we discuss whether these therapeutic obstacles are specific to transcription-factor-mediated reprogramming or inherent to every cellular reprogramming method. Finally, we address whether a better understanding of the mechanism underlying the reprogramming process might improve the fidelity of reprogramming and, therefore, the iPSC quality.
Main Text
In 2005, we were in a lab retreat discussing how many genes would be required to reprogram somatic cells to pluripotency. No one suggested just four proteins. The following year, Takahashi and Yamanaka (2006)) reported the generation of induced pluripotent stem cells (iPSCs) using a reprogramming cocktail of Oct4, Sox2, Klf4, and Myc (hereafter referred to as OSKM), a discovery that has undoubtedly changed the field of regenerative medicine and our understanding of cellular identity. iPSC technology is based on the assumption that a set of transcription factors expressed in embryonic stem cells (ESCs) is responsible for maintaining a pluripotent fate and is sufficient for establishing a de novo pluripotency program. Finding this specific combination of factors is an extraordinary accomplishment considering that ESCs express thousands of proteins. Nicely, the same cocktail of proteins was later shown to also reprogram human somatic cells to pluripotency (Takahashi et al., 2007). The beauty of this finding is that a simple experiment with a very low probability for success answered a complex question. We wonder whether Yamanaka received funding for this specific experiment, considering that many grants are awarded based on the probability of generating the expected results.
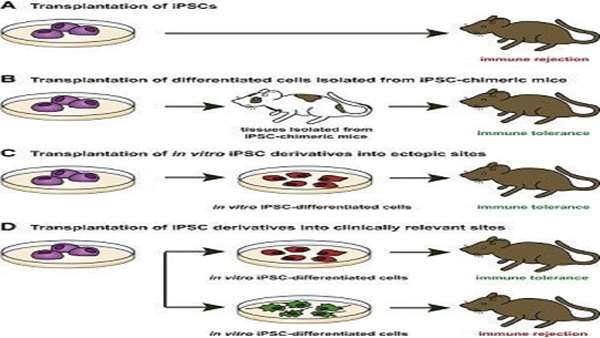
In the last 10 years, iPSCs have been thoroughly scrutinized, and their value as a disease model and a source of cells has been intensively debated. Indeed, genetic mutations and chromosomal aberrations detected in iPSCs have raised concerns about their tumorigenic potential (Yamanaka, 2012). Likewise, epigenetic aberrations have questioned iPSC differentiation potential and immune tolerance after autologous transplantation (Okita et al., 2011). However, further analyses have demonstrated that these abnormalities are mostly due to technical limitations, thus excluding these reprogramming errors as an intrinsic characteristic of transcription factor-mediated reprogramming. Here we review the immunogenic and tumorigenic features attributed to some iPSC clones, the relationship between these undesirable traits and incomplete reprogramming, and the mechanisms underlying different reprogramming methods as a strategy for improving iPSC reprogramming fidelity and thus the utility of iPSCs in molecular and biomedical applications.
Molecular Obstacles to Clinical Translation of iPSCs. Volume 19, Issue 3, p298–309, 1 September 2016. DOI: http://dx.doi.org/10.1016/j.stem.2016.06.017



ارسال به دوستان