The discovery of CRISPR-Cas enzymes in bacteriophages has revealed a new source of potential tools for editing the genome.
Bacteria and archaea use CRISPR-Cas systems to defend themselves from viruses. The repurposing of the systems by humans has transformed genome editing in recent years and spurred interest in finding new CRISPR systems with novel features. The discovery of CRISPR-Cas loci in phage genomes reconstructed from microbial community DNA sequences suggested the viruses, which infect and replicate in bacterial cells, are a potential source of systems that could be repurposed for genome editing.
Nobel Laureate in Chemistry Jennifer Doudna, whose work at the University of California, Berkeley led to the discovery of CRISPR-Cas9 as a tool for making targeted changes to the genome, and her colleagues followed up on the hint that phages could be a source of new systems for genome editing.
Writing in a November 23 article in the journal Cell, the researchers describe a metagenomic analysis of microbial samples isolated from soil, aquatic, human, and animal microbiomes. The comprehensive study of CRISPR-Cas
systems throughout the virosphere revealed "the widespread occurrence of diverse, compact CRISPR-Cas systems encoded in phage genomes." Thousands of viruses encode CRISPR systems, according to the study.
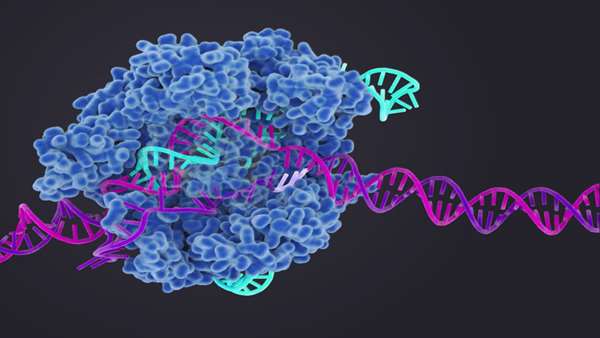
The phage-encoded systems found by Doudna and her collaborators encompass all six known CRISPR-Cas types. Genome-resolved metagenomics and bioinformatics-enabled phylogenetic insights enabled the analysis of the systems from uncultivated viruses and suggested the roles they play.
An investigation of novel hypercompact and divergent CRISPR-Cas systems zeroed in on Casλ. While the team is yet to define some mechanistic aspects of Casλ, the enzyme family has a compact structure that is capable of editing the genomes of plants and humans. The researchers see the enzyme as a "valuable tool" for genome editing.
Further work could unlock the full value of Casλ, with the researchers noting that their "structural understanding" of the enzymes "provides a starting point for the future design of variants." The new variants could have "expanded genome editing capabilities that combine the advantages of a small protein with the versatility of a robust RNA-guided DNA recognition machine."
The findings have caught the attention of researchers in the field. Computational biologist Kira Makarova at the U.S. National Center for Biotechnology Information, who is not an author of the study, explained the implications to the journal Nature.
"This is a significant step forward in the discovery of the enormous diversity of CRISPR-Cas systems. There is a lot of novelty discovered here," said Makarova, who sees plasmids, small circular DNA molecules that transfer DNA between microbes, as another place that scientists will look for CRISPR-Cas systems.



ارسال به دوستان